Basic Syntax
Please select one of the topics for more details.
General Tips for Using R Syntax
When coding in any language, you must first recognize that certain symbols or combinations of symbols correspond to specific actions. Below are some general tips when coding in the R language:
- On any one line of code, anything after a
#symbol will not be executed. This is useful for adding comments and notes to your code or preventing R from executing certain lines without having to delete them from your scripts.
2 + 2
2 + 2 # Ahhh...yeah, this is still 4
# note to self: the expression below returns the value 4
2 + 2
#1 + 2
2 + 2 # 1 + 2 will not be executed, but this line will still return 4- Blank spaces in code are generally ignored and can be used to help separate 'busy' areas of code:
(2+4)/3*5
(2+4) / 3 * 5 # still returns 10, but is easier to read- Both
=and<-have the same meaning when defining R objects:
x = 2
x <- 2- Defining an R object will not return anything in the Console, but calling a defined object will print its value:
x = 2 # x will appear in the Environment window, but not the Console
x # the value 2 will be returned in the Console window- Parentheses,
( ), are used to call functions like:
x <- c(1, 9, 8, 0) # using the combine function, c(), to concatenate the elements
sum(x) # returns 18- Brackets,
[ ], are used to index into a data structure (matrix, data frame, etc.):
x <- c(1, 9, 8, 0)
x[3] # returns the third element, - Most functions have optional arguments that are called within the parentheses of the function:
x <- c(1, 9, NA, 0) # NA represents a missing element
sum(x) # returns NA
sum(x, na.rm = TRUE) # returns 10 because na.rm = TRUE means to remove NAs
Identifying Missing Values
Missing data in R appears as NA. NA is not a string or a numeric value, but an indicator that something is missing. When importing data into R, there can be missing entries, which R fills in with NA. However, it is also possible to create vectors with missing values as shown in the last example above.
If we want to identity which value in a vector is missing, we can use is.na(). Calling this function returns the logical output (i.e., TRUE or FALSE) for each entry in the vector, where TRUE corresponds to the missing entry.
x <- c(1, 9, NA, 0) # NA represents a missing element
is.na(x) # returns FALSE FALSE TRUE FALSE
Data Structures
It is important to know the type and size of an object you are working with since basic operations require objects be of the same type and sometimes the same size. Further, calling upon functions in R requires input objects ('arguments') of the called function to be a certain type and dimension before they can be used.
The five basic data structures used by R are presented in the table below. They are organized by their dimensionality (one-dimensional, two-dimensional, or with n dimensions) and whether they are homogeneous (contents are the same type) or heterogeneous (contents are different types).
Note: There are no scalars in R. Instead, an individual element is considered to be a vector of length one.
There are two general types of data structures in R.
- Homogeneous types, where every element has to be of the same class or is typically of the same class. These include atomic vectors, matrices, and arrays.
- Heterogeneous types, where the elements can be a mix of different classes. These include lists, data frames, and tibbles.
Out of all of these, you will find that lists are the most versatile while data frames, and to some extent tibbles, are most commonly used for managing data in R.
Atomic Vectors
- Four vector types (0:25)
- Functions for identifying object types (1:43)
- Indexing vectors (3:15)
- Combining different vector types (5:20)
- Total length (8:36)
Examples of the four common types of atomic vectors (double, integer, logical, and character) are shown below. Notice that c(), the function for 'concatenate' or 'combine', has been used to create some of the vectors. As shown in the example code, be sure to use commas in between vector elements when calling c().
# Creating different types of atomic vectors
x <- c(134.04, 53.23, 63.39, 116.89) # double (or numeric) vector
y <- 1:4 # integer vector
z <- c(TRUE, FALSE, TRUE, TRUE) # logical vector
stock.sym <- c('IBM', 'MSFT', 'ALL', 'FB') # character vector
Although object type and size can be identified in RStudio’s Environment window, you can also call upon the typeof(), class(), str(), and length() functions directly to provide object information in the Console window.
typeof(x) # returns "double"
class(x) # returns "numeric", which can be more informative
str(x) # returns what is shown in the Environment window
length(x) # returns 4Indexing Atomic Vectors
To refer to a subset of elements from an atomic vector, use brackets after the name of the object. Notice how c() is once again used to call upon the first and third element of the vector, while a colon is used to subset a series of elements.
# Indexing Atomic Vectors
x # returns the entire vector to the Console
x[2] # 2nd element of vector x
x[c(1,3)] # 1st and 3rd elements
x[2:4] # 2nd, 3rd, and 4th elements
Matrices
- Creating a matrix (begins at 0:27)
- Providing row and column names (begins at 2:10)
- How to view contents of a matrix (begins at 2:57)
- Total length (4:28)
Let us now move on to the matrix, which is a two-dimensional data structure similar to atomic vectors. To create a matrix we use the matrix() function. This function requires a data input and typically an input for the number of rows or for the number of columns. Notice in the two examples shown below that the data is directly provided by using the c(), and that the nrow and ncol options require the use of an equals sign.
> matrix(c(2,4,6,8,10,12), nrow=2)
[,1] [,2] [,3]
[1,] 2 6 10
[2,] 4 8 12
> matrix(c(2,4,6,8,10,12), nrow=3)
[,1] [,2]
[1,] 2 8
[2,] 4 10
[3,] 6 12
> matrix(c(2,4,6,8,10,12), ncol =2)
[,1] [,2]
[1,] 2 8
[2,] 4 10
[3,] 6 12You can also call upon matrix() using a predefined vector as the data input to the function.
x <- c(2,4,6,8,10,12)
matrix(x, nrow = 2)- Using the Help menu to go over argument options (begins at 0:16)
- Viewing contents of a matrix and the need for space allocation (begins at 2:11)
- Updating & creating row/column names (begins at 4:11)
- Total length (7:39)
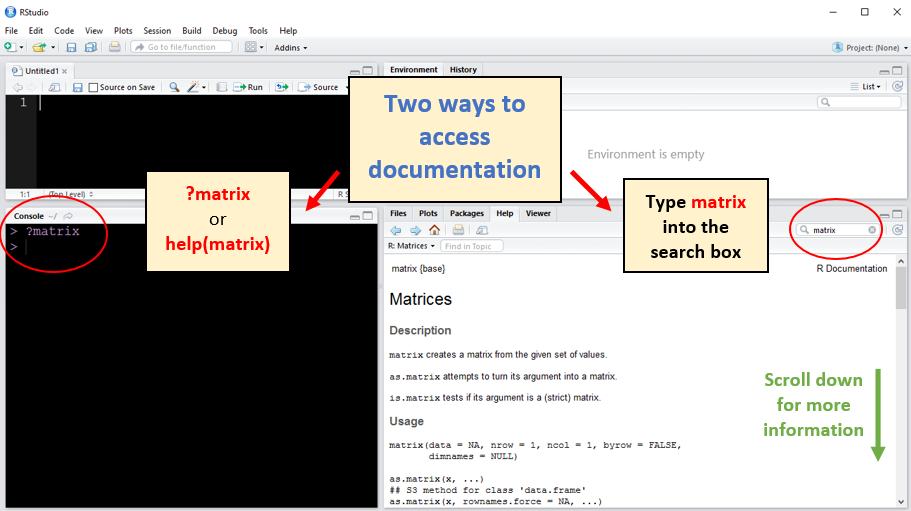
Note: Although we only used two arguments (i.e., inputs) when calling matrix() for this simple example, there are actually more options available; however, not every argument in every function must be specified. An online search can provide details for nearly every R function, but there are more convenient ways of accessing this information from within RStudio. We can type ?matrix or help(matrix) into the Console or use the search box at the top of RStudio’s Help window.
- Defining of example data (begins at 0:27)
- Using
rbind()to add rows to a matrix (begins at 0:47) - Indexing into a matrix (begins at 1:50)
colSums(),colMeans(),rowSums(), androwMeans()(begins at 3:40)- Simple arithmetic operations (begins at 4:15)
- Using
cbind()to add columns to a matrix (begins at 5:40) - Defining nominal variables (begins at 6:12)
- Defining ordinal variables (begins at 8:12)
- Total length (10:12)
Note: Similar to vectors, matrices can be indexed into and used in simple arithmetic operations. However, there are other functions such as colSums() and colMeans() that can make matrices a better data structure for some storage needs.
Data Frames
- Creating a data frame (begins at 0:25)
- Comments on row and column names (begins at 1:30)
- Functions useful for retrieving information (begins at 2:35)
- Indexing into a data frame (begins at 4:00)
- Total length (8:25)
Though matrices and vectors are common structures in programming, most datasets in R are actually data frames. Data frames allow an analyst to more efficiently manage information by taking advantage of what goes on behind the scenes when code is being processed. To create a data frame we use the data.frame() function.
Consider the example shown below, where we would like to compare quarterly stock prices for eight different stocks using the following information:
- names – stock symbols for the eight companies
- Q1 – first quarter stock price
- Q2 – second quarter stock price
- Market – market indicator
- Risk – ordinal variable created for this example by ranking each of the eight companies as having a low, medium, or high level of volatility relative to each other
# Define our components
names <- c('IBM', 'MSFT', 'ALL', 'FB', 'AMZN', 'AAPL', 'GOOG', 'PGR')
Q1 <- c(134.04, 53.23, 63.39, 116.89, 577.72, 100.22, 728.56, 32.47)
Q2 <- c(150.02, 51.23, 67.29, 111.08, 699.33, 96.21, 706.94, 31.13)
Market <- c('NYSE', 'NASDAQ', 'NYSE', 'NASDAQ', 'NASDAQ', 'NASDAQ', 'NASDAQ', 'NYSE')
Risk <- factor(c('Low', 'Medium', 'Medium', 'Low', 'High', 'High', 'Medium', 'Low'),
order = T, levels = c('Low', 'Medium', 'High'))# Combine all into a single data frame
stocks_df <- data.frame(row.names = 1, names, Q1, Q2, Market, Risk)Notice that the last line of code creates the data frame stocks_df using data.frame() with the added argument of row.names. This means that names will not be recognized as a variable in the dataset. This is done because names only serves as an identifier variable, which can be used to help index into the data frame for subsets of information. The remaining inputs after the row.names argument simply state the four column vectors which make up our example dataset.
Note: One advantage of creating a data frame structure is that character vectors (such as Market in the example above) are automatically recognized as categorical variables. To recreate this as a matrix structure would take additional lines of code, which consequentially could result in unforeseen errors. Also, remember that you can type ?data.frame into your Console to bring up the RStudio’s help documentation for further details about how to use this powerful function.
The following are useful functions for gaining general knowledge about a dataset and for taking a peak at the contents:
dim(stocks_df) # checks the dimensions - rows (observations) by column(variables)
head(stocks_df) # displays the first 6 rows for all variables
str(stocks_df) # structure - same info as shown in the Environment window
summary(stocks_df) # summary by variable (depends on variable type)Indexing a Data Frame
As shown in the example lines of code below, creating row names helps with indexing into a data frame to extract specific content. Again, this can be done with matrices; however it would require more lines of code. Further, only with a data frame or list structure can you use the $ syntax as shown in the last line of code below to easily index a specific column of information.
## Indexing the data frame stocks_df
stocks_df[1, 2] # 1st row, 2nd column (i.e., Q2 for IBM)
stocks_df['IBM', 'Q2'] # same as above
stocks_df[6, ] # All variables for Apple
stocks_df['AAPL', ] # same as above
stocks_df[3:7, 'Risk'] # Risk level for company stocks 3 through 7
stocks_df$Risk[3:7] # same as aboveAnd as a final illustration, you can also use conditional statements in combination with the above syntax.
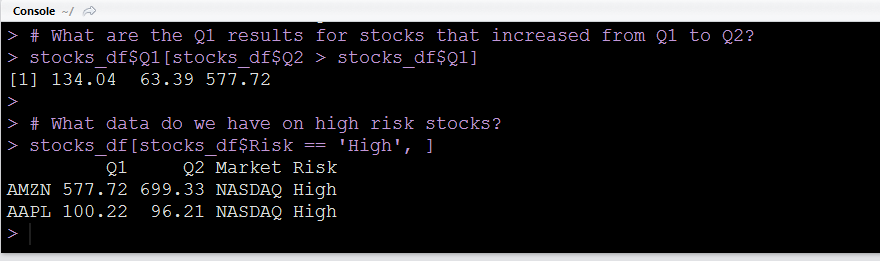
Note: You could also use subset() as a more compact form of the last line of code above
# What data do we have on high risk stocks?
subset(stocks_df, Risk == 'High')
Lists
Lists are fundamentally different from other data structures because their components can be of different lengths and types. Let's first create some example components and then create a list using the list()function.
# Creating different data structure components
v <- c('Moe', 'Larry', 'Curly') # character vector
m <- matrix(c(1:6), nrow = 2) # matrix
df <- data.frame( Who = c('Bill', 'Sue'), age = c(19, 23)) # data frame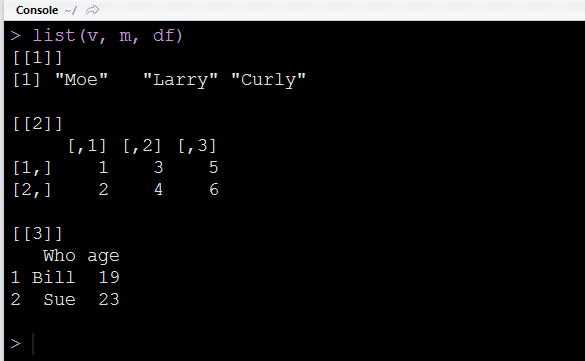
Notice that we only had to separate each component with a comma when using list().
Indexing a list
When we refer to the Console output above, we can see that R is trying to tell us that we can use double brackets, [[ ]], to index into a list.
L1 <- list(v, m, df) # define list L1
L1[[1]] # "Moe" "Larry" "Curly"
L1[[1]][2] # "Larry"
L1[[3]]$Who # Bill Sue
L1[[3]]$age[2] # returns the value 23Note: In the last two lines of code, we could use the $ syntax to index the data frame because we named the components inside the data frame beforehand. This approach can be extended to lists as well.
L1 <- list(stooges = v, mat = m, results = df) # define list L1 using names
L1$stooges # "Moe" "Larry" "Curly"
L1$stooges[2] # "Larry"
L1$results$Who # Bill Sue
L1$results$age[2] # 23
Combing Matrices and Atomic Vectors
Sometimes we need to combine matrices and/or vectors into one matrix. This can be done by using the rbind() and cbind() functions. Take the following example where we wish to bind row bto matrix A.

This can be accomplished by using rbind() since we are binding rows.
A <- matrix(c(2,4,6,8,10,12), ncol = 3) # defining A as a 2-by-3 matrix
b <- c(20, 30, 40) # defining b as a 1-by-3 row vector
rbind(A, b) # row binding to form a 3-by-3 matrixNotice that the number of columns for A and b had to match. Otherwise, R would return an error. Similarly, we could use cbind() to bind together column vectors to form a matrix, or many matrices to form one larger matrix, given that their number of rows were the same.